|
Site directed engineering of phytase B for activity at varied pH.
Vineet Agrawal and Nilanjan Roy
The enzyme, phytase hydrolyzes phytic acid myo-inositol hexakisphosphate the primary storage form of phosphate in cereals
and oilseeds, to myo-inositol and phosphoric acid. It’s an essential animal feed adjuvant especially in monogastric
animals having no phytase activity to increase phosphate bioavailiability and reduce phosophorus pollution. However the use
is limited by low availiability, restricted pH dependent activity (pH 2.5 or pH 5.0) & low thermostabilty. An overexpressed
modified phytase can thus be of immense value commercially. On the basis of this Phytase B from Aspergillus niger var
Teigham was cloned and expressed in Saccharomyces cervisiae. The sequence analysis demonstrated 90% identity with a
highly conserved catalytic site and a substituted Serine (D75S) in the substrate specificity site. The electrostatic potential
calculated using MOE illustrated a positive catalytic region at both pH. However, substrate specificity site demonstrate pH
dependent positive or negative region at pH 2.5 or 5.0 respectively. A positive electrostatic region at pH 5.0 can enhance
the binding of highly negatively charged substrate phytate. The identification of such amino acid residues and subsequent
replacement with site directed mutagenesis to yield a wide pH range active phytase B is in progress.
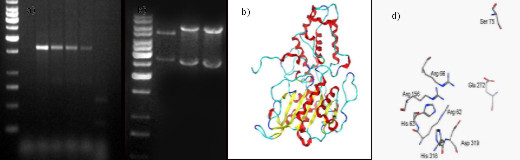
phytase gene was amplified by PCR (a) cloned in expression vector (b) Homology modeled & (c) active site residues identified
(d)
Cloning and characterization of oxidoreductase from
Geotrichum candidum.
Sonia Pahwa and Nilanjan Roy
Geotrichum oxidoreductases play important role in various biocatalytic reactions. Aromatic and aliphatic racemic alcohols
can be converted to the corresponding optically active alcohols with excellent enantio-selectivity and high yield using Geotrichum
candidum oxidoreductases. Here, we report the cloning and characterization of an oxidoreductase, from Geotrichum. Genomic
library was constructed in pRS314 and an oxidoreductase is cloned by functional complementation of yeast alcohol dehydrogenase
(ADH) mutant. After sequencing the gene is identified as histidinol dehydrogenase. L-histidinol-NAD oxidoreductase (EC1.1.1.23)
catalyzes final two steps in the biosynthesis of the amino acid L-histidine. The deduced amino acid sequence (327 amino acids)
displayed 89% similarity with bacterial histidinol dehydrogenas and 96% similarity with Pichia patoris and S. cerevisiae
phosphoribosyl-AMP cyclohydrolase. We also found that Geotrichum HDH gene can complement S. cerevisae adh deletion
mutants in vivo and can take ethanol as substrate in vitro.
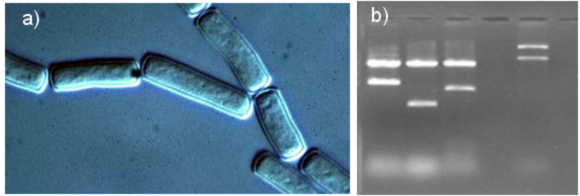
Histidinol dehydrogenase gene (b) is cloned from G. candidum (a)
|
